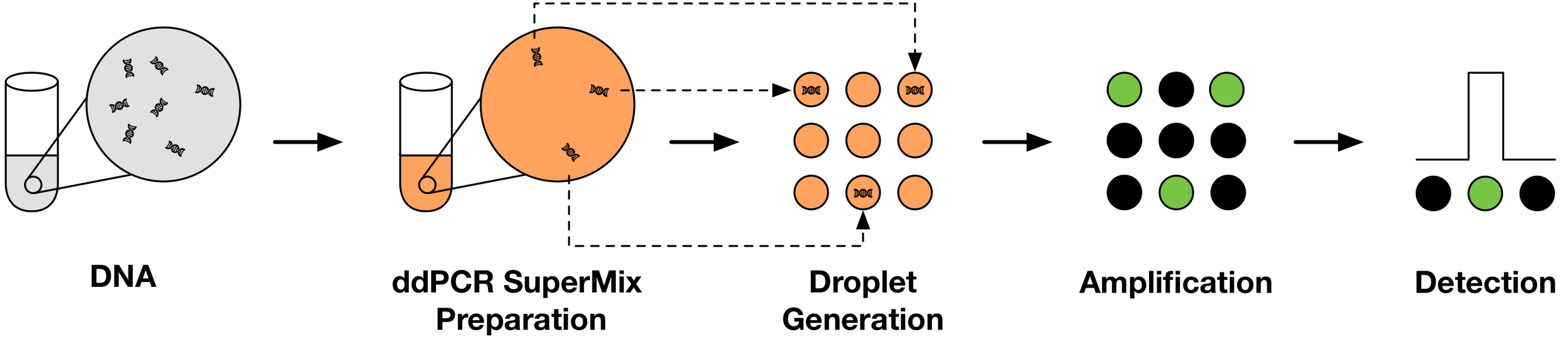
Single-Copy Gene Quantification Using ddPCR
Droplet digital polymerase chain reaction (ddPCR) partitions a sample into roughly 20,000 water-oil emulsion droplets to create a massively parallel number of PCR reactions. Droplets containing an amplified product are then measured as highly fluorescent due to the presence of a DNA-specific fluorescent dye. When a solution is adequately dilute, each droplet will contain either zero or one copy of a gene sequence allowing us to accurately quantify genome copies without the need for standards.
Mojarro et al. (2017) LPS XLVIII ePoster
This technique was implemented in Mojarro et al. (2019) Astrobiology, PDF in order to validate the extraction of 70 pg of DNA.
Bacillus subtilis (ATCC 6633) Spore Genome Quantification Protocol
Since one B. subtilis spore contains only one copy of its genome, we can utilize a single-copy primer to count individual spores. Starting spore concentrations are quantified by colony counting on LB plates.
SpaC gene:
Forward primer: TGAGGAAGGATGGGACGACA
Reverse primer: AACAGATTGCTGCCAGTCCA
ddPCR Reaction Mixture:
Template DNA - 5 µL
Water (balance) - 3 µL
ddPCR Evagreeen Supermix (2x) - 10 µL
Forward Primer (3.3 µM) - 1 µL
Reverse Primer (3.3 µM) - 1 µL
Total Reaction - 20 µL
Thermocycler Conditions for Bio-Rad C1000 (from QX200 ddPCR EvaGreen Supermix pdf):
*use heated lid set to 105 C, sample volume to 40 µL, and ramp rate to 2 C/sec for all steps.
Enzyme activation @ 95º C for 5 minutes
Denaturation @ 95º C for 30 seconds
Annealing/extension @ 60º C for 1 minute
Repeat steps 2-3 40 times
Signal stabilization @ 4º C for 5 minutes
Signal stabilization @ 90º C for 5 minutes
Hold (optional) @ 4º C until ready to measure
SpaC primers and B. subtilis DNA
Example of raw ddPCR data illustrating detection of individual spores (DNA copies relative fluorescence greater than 10,000).
The baseline fluorescence is determined by primer-dimer products.
Escherichia coli (OP50) Vegetative Cell Genome Quantification Protocol
Accurately quantifying DNA yield from vegetative E. coli cells is a bit more complicated. Depending on your culture's growth stage one E. coli cell can have 1 - 3 copies of its genome. Therefore, we must supplement our OD 600 measurement with genome copy information to correct for the growth stage. Otherwise it is very easy to over/under estimate your DNA yield from vegetative cells. Here we utilize a single-copy E. coli primer to discriminate genome copies.
MetG gene:
Forward primer: GGTGGAAGCCTCTAAAGAAGAAG
Reverse primer: AGCAGTTTGTCAGAACCTTCAAC
Escherichia coli Genome Copy Number Correction Protocol
Take the OD 600 measurement after n hours from your initial inoculation.
Ex. 1.8 x 10⁸ cells/mL
Dilute/wash your cells in phosphate-buffered saline solution for colony ddPCR. Remember to take note of the ddPCR detection range.
Ex. 180 cells/µL
Pipet 5 µL of your washed/diluted cells directly into your ddPCR reaction mixture in place of the "template DNA" volume. Use the same reaction mixture and thermocycling parameters as above.
Calculate genome copies per cells using your OD 600 measurements and ddPCR results.
genome copies/cell = measured cells into ddPCR / ddPCR genome copies
Ex. 2 copies/cell = 1800 genome copies / 900 cells